Cancer Epigenetics Lab (CEL)
The Cancer Epigenetics Lab (CEL) hosts two independent groups. Karim Malik’s group currently focuses on (i) delineating cellular functions in development and cancer of protocadherins (recently shown to be epigenetically silenced in cancer), (ii) evaluating novel drugs for the treatment of neuroblastoma, and (iii) transcriptional regulation of, and by, WT1. Together with Keith Brown’s group, we have conducted genome-wide epigenetic profiling of Wilms tumour and neuroblastoma. Keith Brown’s group also works on genetic-epigenetic interactions regulated by the neuroblastoma oncogene NMYC.
The CEL, previously the CLIC Sargent Research Unit, was set up in 1985 to study the fundamental changes that cause cancers to develop in children. During that time many exciting discoveries have been made around the world, and the CEL has made important contributions to this increased understanding of cancer. This work is vital to ensure that more children are cured of their cancers, and to improve on the treatments already in use, so that unpleasant side effects can be avoided.
The changes that lead to cancers are defects in the genetic information, which is present in our DNA. The DNA constitutes a sort of library of plans, with each book representing a gene. There are about 30,000 human genes. Each gene codes for the production of a specific protein that performs a function in the cell, such as providing a supporting structure, an enzyme that makes an essential chemical, or a controlling factor that tells cells when to grow. Cells need this genetic information to tell them how to replicate themselves and perform their day-to-day functions. Genes can become defective either because the DNA becomes damaged (mutated), or because too much or too little protein is made from them. If defects occur in a gene that tells a cell how and when to divide, then it can start to grow uncontrollably, and so develop into a tumour. Identifying the exact genes that may become altered during the development of cancer is a major aim of much current research, including that of the CEL. Recently we have concentrated a tumour of the kidney, called Wilms' tumour, and another childhood cancer, neuroblastoma.
- Our short-term aims are to identify the genes involved in childhood tumours and to how they cause cancer when they go wrong.
- Our long-term aims are to translate this knowledge into new rational therapies for childhood cancer.
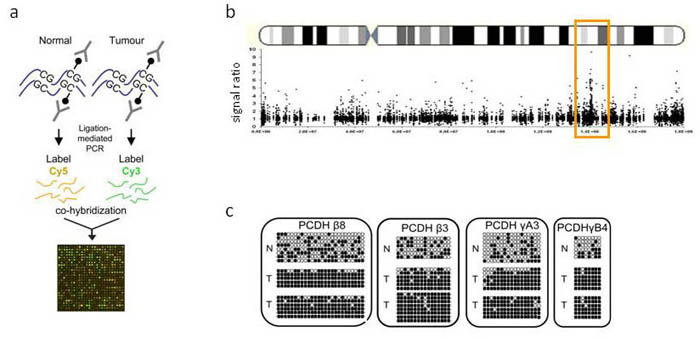
Long range epigenetic silencing in Wilms' tumour: (a) Methylated DNA from normal kidney and Wilms' tumour was captured by Methyl-DNA immunoprecipitation (MeDIP), then fractions were fluorescently labelled and hybridised to promoter microarrays, (b) The averaged MeDIP-chip profile of several Wilms' tumours showed a peak of DNA methylation on the long arm of chromosome 5, which contained the protocadherin (PCDH) α, β and γ clusters,(c) DNA methylation assayed using bisulphite sequencing for protocadherin genes mapped within the methylated region. White circles represent unmethylated CpG residues and filled circles methylated CpGs. Tumours (T) are hypermethylated compared to normal tissue (N).
Our work has shown that there are many different genes that can cause Wilms' tumour when defective i.e. there are several routes to the same kind of tumour, and that there are several different sorts of changes by which these genes become defective. In particular, we have concentrated on epigenetic alterations, which are caused by subtle changes that alter the activity of genes so that too much or too little protein in made. We are studying epigenetic changes in genes that we already know are important in Wilms' tumour, as well as other new genes that we have found. One of the important aspects of epigenetic aberrations is that they are potentially reversible, unlike other alterations in cancer. Thus studies of epigenetic changes could lead to completely new ways of treating cancer.
Selected Publications
- Charlet, J., Tomari, A., Dallosso, A.R., Szemes, M., Kaselova, M., Curry, T.J., Almutairi, B., Etchevers, H., McConville, C., Malik, K. and Brown, K. (2016) Genome-wide DNA methylation analysis identifies MEGF10 as a novel epigenetically repressed candidate tumour suppressor gene in neuroblastoma. Molecular Carcinogenesis E-pub ahead of print
- Vieira, G.C., Sankarakuttalam, C., Melegh, Z., Greenhough, A., Malik, S., Szemes, M., Park, J.H., Kaidi, A., Zhou, L., Catchpoole, D., Morgan, R., Bates, D.O., Gabb, P.D. and Malik, K. (2015) LGR5 regulates pro-survival MEK/ERK and proliferative Wnt/β-catenin signalling in neuroblastoma. Oncotarget 6, 40053-40067.
- Park, J.H., Szemes, M., Vieira, G.C.C., Melegh, Z.B., Malik, S., Heesom, K.J., Von Wallwitz-Freitas, L., Greenhough, A., Brown, K.W., Zheng, Y.G., Catchpoole, D., Deery, M.J. and Malik, K.T.A. (2015) Protein arginine methyltransferase 5 is a key regulator of the MYCN oncoprotein in neuroblastoma cells. Molecular Oncology 9, 617-627.
- Charlet, J., Szemes, M., Malik, K.T.A. and Brown, K.W. (2014) MYCN is recruited to the RASSF1A promoter but is not critical for DNA hypermethylation in neuroblastoma. Molecular Carcinogenesis 53, 413-420 MC-12-0258.R1
- Szemes, M., Dallosso, A.R., Melegh, Z., Curry, T., Li, Y., Rivers, C., Uney, J., Mägdefrau, A., Schwiderski, K., Park, J., Brown, K.W., Shandilya, J., Roberts, S.G.E. and Malik, K. (2012) Control of epigenetic states by WT1 via regulation of de novo DNA methyltransferase 3A. Human Molecular Genetics 22, 74-83.
- Brown, K.W., Charles, A., Dallosso, A.R., White, G., Charlet, J., Standen, G.R. and Malik, K. (2012) Characterization of 17.94, a novel anaplastic Wilms' tumor cell line. Cancer Genetics and Cytogenetics, 205, 319-326.
- Dallosso, A.R., Øster, B., Greenhough, A., Thorsen, K., Curry, T.J., Owen, C., Hancock, A.L., Szemes, M., Paraskeva, C., Frank, M., Andersen, C.L. and Malik, K. (2012) Long-range epigenetic silencing of chromosome 5q31 protocadherins is involved in early and late stages of colorectal tumorigenesis through modulation of oncogenic pathways. Oncogene Advanced online publication 16 Jan 2012.
- Kim, M.S., Yoon, S.K., Bollig, F., Kitagaki, J., Hur, W., Whye, N.J., Wu, Y.P., Rivera, M.N., Park, J.Y., Kim, H.S., Malik, K., Bell, D.W., Englert, C., Perantoni, A.O. and Lee, S.B. (2010). A novel Wilms Tumor 1 (WT1) target gene negatively regulates the WNT signaling pathway. J. Biol. Chem. 285(19), 14585-14593.
- Dallosso, A.R., Hancock, A.L., Szemes, A., Moorwood, K., Chilukamarri, L., Tsai, H-H., Sarkar, A., Barasch, J., Vuononvirta, R., Jones, C., Pritchard-Jones, K.m Royer-Pokora, B., Lee, S.B., Owen, C., Malik, S., Feng, Y., Frank, M., Ward, A., Brown, K.W. and Malik, K. (2009) Frequent long-range epigenetic silencing of protocadherin gene clusters on chromosome 5q31 in Wilms' Tumor. PLoS Genetics, 5(11), e1000745 [Available online].