Gene expression lab
Our aim is to understand how transcriptional regulatory proteins function. To achieve this goal we are combining the study of general mechanisms with the function of a specific and complex transcriptional regulator, the Wilms’ tumour suppressor protein WT1.
General transcription mechanisms
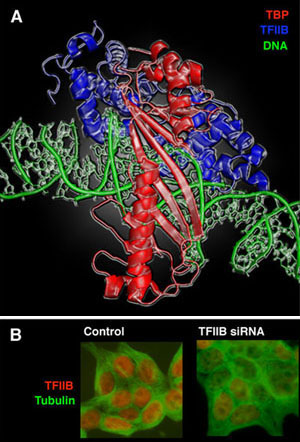
The general transcription factor TFIIB plays a central role in the assembly of the transcription complex at the gene promoter. TFIIB is also a target of transcriptional activator proteins, which act to increase transcription complex assembly. We have been investigating these integrated roles of TFIIB and have uncovered two novel regulatory features of TFIIB that are critical to its function. Firstly, TFIIB possesses extensive DNA-binding ability, which we have shown to play a novel and critical role in transcriptional regulation. Secondly, our studies provided the first insights into the essential role of TFIIB conformation in transcriptional regulation.
We are currently exploiting an RNAi-based system to analyze TFIIB derivatives in mammalian cells in the absence of endogenous TFIIB (Figure 1). This cell-based approach is complemented by biochemical analyses, which together are employed to analyze critical functions of TFIIB in transcription initiation including; promoter recognition, transcription initiation control and the mechanisms of transcriptional activation.
The mechanism of action of the Wilms' tumour suppressor protein WT1
Since its discovery WT1 has proved to be a fascinating protein that has roles both in transcriptional regulation and RNA processing. WT1 was originally identified as a gene product that is frequently mutated or abnormally expressed in Wilms’ tumours, the most common solid childhood malignancy. However, WT1 has since been found to play a role in several other malignancies, including leukemia, breast and lung cancer. Our interests concern the mechanisms by which WT1 regulates transcription and it’s role in both development and disease.
We discovered that the transcriptional activity of WT1 is regulated by interaction with a cosuppressor (Figure 2). We have identified one of the components of the cosuppressor complex (BASP1) and are currently in the process of identifying the remaining factors. BASP1 also exhibits tumour suppressor activity and its intracellular distribution can be regulated by myristoylation, phosphorylation and sumoylation. How these modifications regulate the WT1-BASP1 dynamic and their role in transcriptional regulation by WT1 in differentiation and disease are a current focus of our work.

Selected Publications
- Shandilya, J., Wang, Y. and Roberts,S.G.E. (2012). TFIIB dephosphorylation links transcription inhibition with the p53-dependent DNA damage response. Proceedings of the National Academy of Sciences USA 109, 18797-18802.
- Toska, E., Campbell, H.A., Shandilya, J., Goodfellow, S.J., Shore, P., Medler, K.F. and Roberts, S.G.E. (2012) Repression of transcription by WT1-BASP1 requires the myristoylation of BASP1 and the PIP2-dependent recruitment of histone deacetylase. Cell Reports 2, 462-469.
- Essafi, A., Webb, A., Berry, R.L., Slight, J., Burn, S.F., Spraggon, L., Velecela, V., Martinez-Estrada, O.M., Wiltshire, J.H., Roberts, S.G.E., Davies, J.A., Hastie, N.D. and Hohenstein, P. (2011) A Wt1-controlled chromatin switching mechanism underpins tissue-specific Wnt4 activation and repression. Developmental Cell 21, 559-574.
- Goodfellow, S.J., Rebello, M.R., Zeef, L.A.H., Toska, E., Rudd, S.G., Medler, K.F. and Roberts, S.G.E. (2011) WT1 and its transcriptional cofactor BASP1 redirect the differentiation pathway of an established blood cell line. Biochemical Journal 435, 113-125.
- Hartkamp, J., Carpenter, B. and Roberts, S.G.E. (2010) The Wilms' Tumour Suppressor Protein WT1 is processed by the Serine Protease HtrA2/Omi. Molecular Cell 37, 159-171.
- Wang. Y., Fairley, J.F. and Roberts, S.G.E. (2010) Phosphorylation of TFIIB links transcription initiation and termination. Current Biology 20, 548-553.
- Deng, W., Malecová, B., Oelgeschläger, T. and Roberts, S.G.E. (2009). TFIIB recognition elements control the TFIIA-NC2 axis in transcriptional regulation. Molecular and Cellular Biology 29, 1389-1400.
- Green, L.M., Wagner, K.J., Campbell, H.A., Addison, K. and Roberts, S.G E. (2009) Dynamic interaction between WT1 and BASP1 in transcriptional regulation during differentiation. Nucleic Acids Research 37, 431-440.
- Deng, W. and Roberts, S.G.E. (2007) TFIIB and the regulation of transcription by RNA polymerase II. Chromosoma 116, 417-429.
- Elsby, L. M., O’Donnell, A. J. M., Green, L. M., Sharrocks, A. D. and Roberts S. G. E. (2006) Assembly of TFIIB at a promoter in vivo requires contact with RNA polymerase II. EMBO Reports 7, 898-903.
- Deng, W. and Roberts, S.G.E. (2005) A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes and Development 19, 2418-2423.
- Roberts, S.G.E. (2005) Transcriptional regulation by WT1 in development. Current Opinion in Genetics and Development 15, 542-547.
- Carpenter, B., Hill, K.J., Charalambous, M., Wagner, K.J., Lahiri, D., James, D.I., Anderson, J.S., Schumacher, V., Royer-Pokora, B., Mann, M., Ward, A. and Roberts, S.G.E. (2004) BASP1 is a transcriptional cosuppressor for the Wilms’ tumour suppressor protein WT1. Molecular and Cellular Biology 24, 537-549.
- Glossop, J.A., Dafforn, T.R. and Roberts, S.G.E. (2004) A conformational change in TFIIB is required for activator-mediated assembly of the preinitiation complex. Nucleic Acids Research 32, 1829-1835.
- Schumacher, V., Schuhen, S., Sonner, S., Weirich, A., Leuschner, I., Harms, D., Licht, J., Roberts, S. and Royer-Pokora, B. (2003) Two Molecular Subgroups of Wilms’ Tumors with or without WT1 mutations. Clinical Cancer Research 9, 2005-2014.
- Fairley, J.A., Evans, R., Hawkes, N.A. and Roberts, S.G.E. (2002) Core promoter-dependent TFIIB conformation and a role for TFIIB conformation in transcription start site selection. Molecular and Cellular Biology 22, 6697-6705.
- Richard, D.J., Schumacher, V., Royer-Pokora, B. and Roberts, S.G.E. (2001) Par 4 is a coactivator for a splice-isoform specific transcriptional activation domain in WT1. Genes and Development 15, 328-339.
- Evans, R., Fairley, J.A. and Roberts, S.G.E. (2001) Activator-mediated disruption of sequence-specific DNA contacts by the general transcription factor TFIIB. Genes and Development 15, 2945-2949.
- Hawkes, N.A., Evans, R. and Roberts, S.G.E. (2000) The conformation of the transcription factor TFIIB modulates the response to transcriptional activators in vivo. Current Biology 10, 273-276.